Expressions and Keys
Mathematical and Logical Expressions
Neper handles mathematical expressions thanks to the muparser library. The expression must contain no space, tabulation or new-line characters, and match the following syntax. [1]
Functions
The following table gives an overview of the functions supported by the default implementation. It lists the function names, the number of arguments and a brief description.
Name |
Description |
|
sine function |
|
cosine function |
|
tangens function |
|
arcus sine function |
|
arcus cosine function |
|
arcus tangens function |
|
hyperbolic sine function |
|
hyperbolic cosine |
|
hyperbolic tangens function |
|
hyperbolic arcus sine function |
|
hyperbolic arcus tangens function |
|
hyperbolic arcur tangens function |
|
logarithm to the base 2 |
|
logarithm to the base 10 |
|
logarithm to the base 10 |
|
logarithm to base \(e\) (2.71828…) |
|
e raised to the power of x |
|
square root of a value |
|
sign function: -1 if \(x<0\); 1 if \(x>0\) |
|
round to nearest integer |
|
absolute value |
|
min of all arguments |
|
max of all arguments |
|
sum of all arguments |
|
mean value of all arguments |
Binary Operators
The following table lists the default binary operators supported by the parser.
Operator |
Description |
Priority |
|
logical and |
1 |
|
logical or |
2 |
|
less or equal |
4 |
|
greater or equal |
4 |
|
not equal |
4 |
|
equal |
4 |
|
greater than |
4 |
|
less than |
4 |
|
addition |
5 |
|
subtraction |
5 |
|
multiplication |
6 |
|
division |
6 |
|
raise x to the power of y |
7 |
Ternary Operators
The parser has built in support for the if-then-else operator. It uses lazy evaluation in order to make sure only the necessary branch of the expression is evaluated.
Operator |
Description |
|
if-then-else operator, following the C/C++ syntax: |
Statistical Distributions
The following table lists the statistical distributions. Custom endpoints (not indicated) can also be added as arguments, as described in the following.
Operator |
Description |
Information |
|
normal |
|
|
lognormal |
|
|
Dirac |
|
|
beta function |
\(x>0\), \(y>0\) |
|
Lorentzian |
|
|
Student’s t |
|
|
Weibull |
\(k > 0\) represents the shape |
|
Breit-Wigner |
\(<gamma> \geq 0\), default 1 |
|
exp-normal |
\(<gamma> > 0\), default \(<sigma>\) |
|
Moffat |
\(<gamma> > 0\), default 1 |
|
Pearson type VII |
default \(<gamma> = 1.5\) |
|
Pseudo-Voigt |
\(<gamma> \in [0,\,1]\), default 0.5 |
|
skewed normal |
default \(<gamma> = <sigma>\) |
|
custom |
<mean> represents the mean (or centre), and <sigma> represents the standard deviation (or scale, \(> 0\)). <gamma> depends on the distribution function (see the above table). For all distributions but weibull and beta, custom endpoints can be added as last arguments, as <from_value>,<to_value>, where <from_value> is the lower endpoint and <to_value> is the upper endpoint. The parameter keywords do not need to be provided, but, when they are, the parameters can be given in any order, as in moffat(gamma=1,from=0,to=1,sigma=0.1,mean=0.5). Endpoints are considered inclusive by default, but exclusive endpoints can be specified using fromexclusive=<from_value> and toexclusive=<to_value> (frominclusive=<from_value> and toinclusive=<to_value> can be used for inclusive endpoints). String completion is available for the keywords. Finally, a sum of distributions of increasing averages can be provided, as in 0.3*lognormal(0.5,0.1)+0.7*normal(1,0.1).
Attention
When from and/or to are used, they should preferably be so that the distribution retains the same mean; otherwise, the distribution is shifted after truncation to match the specified mean.
In the case of the custom distribution, the numerical distribution must be provided in the file. The file must contain the x and y values of the distribution on successive lines. The x values must be provided in ascending order and form a regular grid. The distribution must contain at least 3 points and does not need to integrate to 1.
Tessellation Keys
Available keys for a tessellation itself are provided below.
Key |
Descriptor |
Apply to |
|
dimension |
tess |
|
number of vertices |
tess |
|
number of edges |
tess |
|
number of faces |
tess |
|
number of polyhedra |
tess |
|
number of cells |
tess |
|
x coordinate |
tess |
|
y coordinate |
tess |
|
z coordinate |
tess |
|
x, y and z coordinates |
tess |
|
surface area |
tess |
|
volume |
tess |
|
size (surface area/volume in 2D/3D) |
tess |
|
simulation step |
tess |
Available keys for tessellation seeds, vertices, edges, faces, polyhedra, crystals and cell groups are provided below. Also note that the keys apply to cells if they are tagged to apply to polyhedra and the tessellation is 3D and faces and the tessellation is 2D, and that keys apply to crystals if they apply to cells. You may also replace, in the tessellation keys themselves, poly by cell if the tessellation is 3D and face by cell if the tessellation is 2D (it applies only in rare cases). For example, for a 2D tessellation, you may use -statcell ncells instead of -statface nfaces. Keys specific to cells are defined accordingly in the following but also apply to polys is the tessellation is 3D and faces is the tessellations is 2D.
To turn a key value into a value relative to the mean over all entities (e.g. the relative cell size), append the key expression with the :rel modifier. To turn a key value into a value which holds for a unit cell size, append the key expression with the :uc modifier. To use as a reference only the body entities (see below), append b to the modifiers.
Key |
Descriptor |
Apply to |
|
identifier |
seed, ver, edge, face, poly, group |
|
x coordinate |
seed, ver, edge, face, poly |
|
y coordinate |
seed, ver, edge, face, poly |
|
z coordinate |
seed, ver, edge, face, poly |
|
x, y and z coordinates |
seed, ver, edge, face, poly |
|
minimum x coordinate |
edge, face, poly |
|
minimum y coordinate |
edge, face, poly |
|
minimum z coordinate |
edge, face, poly |
|
maximum x coordinate |
edge, face, poly |
|
maximum y coordinate |
edge, face, poly |
|
maximum z coordinate |
edge, face, poly |
|
weight (width for a lamellar tessellation) |
seed, cell |
|
body level |
ver, edge, face, poly |
|
state |
ver, edge, face, poly |
|
type of domain (0 if on a domain vertex, 1 if on a domain edge and 2 if on a domain face) |
ver, edge, face |
|
domain face (-1 if undefined) |
face |
|
domain edge (-1 if undefined) |
edge |
|
domain vertex (-1 if undefined) |
ver |
|
scale |
ver, edge, face, poly, cell [2] |
|
length |
edge |
|
directional length along \((d_x,d_y,d_z)\) (in 2D, |
edge, face, poly |
|
surface area |
face, poly, group |
|
volume |
poly, group |
|
size (surface area/volume in 2D/3D) |
cell, group |
|
equivalent diameter [4] |
face, poly |
|
average equivalent diameter [4] |
face, poly |
|
equivalent radius (half of the eq. diameter) |
face, poly |
|
average equivalent radius (half of the eq. diameter) |
face, poly |
|
sphericity [5] |
poly |
|
circularity [6] |
face |
|
convexity [7] |
face (only for a 2D tessellation), poly |
|
average dihedral angle |
face, poly |
|
minimum dihedral angle |
face, poly |
|
maximum dihedral angle |
face, poly |
|
dihedral angles |
face, poly |
|
flatness fault (in degrees) |
face |
|
disorientation angle (in degrees) |
edge (in 2D), face (in 3D) |
|
cylinder polygonization [19] |
edge |
|
number of vertices |
edge, face, poly |
|
vertices |
edge, face, poly |
|
number of edges |
ver, face, poly |
|
edges |
ver, face, poly |
|
number of faces |
ver, edge, poly |
|
faces |
ver, edge, poly |
|
number of polyhedra |
ver, edge, face |
|
polyhedra |
ver, edge, face |
|
number of neighboring faces |
face |
|
neighboring faces |
face |
|
number of neighboring faces of the same domain (parent cell of a multiscale tessellation) |
face (in 2D) |
|
neighboring faces of the same domain (parent cell of a multiscale tessellation) |
face (in 2D) |
|
number of neighboring polyhedra |
poly |
|
neighboring polyhedra |
poly |
|
neighboring polyhedra, unsorted list |
poly |
|
number of neighboring polyhedra of the same domain (parent cell of a multiscale tessellation) |
poly |
|
neighboring polyhedra of the same domain (parent cell of a multiscale tessellation) |
poly |
|
vertex coordinates |
face, poly |
|
face surface areas |
poly |
|
face equations [9] |
poly |
|
number of neighboring seeds |
poly |
|
neighboring seeds [10] |
poly |
|
identifier of the corresponding cell at scale |
cell |
|
lamella width id [11] |
cell |
|
mode [12] |
cell |
|
group |
cell |
|
periodic (1 if periodic, 0 otherwise) |
ver, edge, face (in 3D) |
|
1 if in orientation fiber and 0 otherwise, see Orientation Fibers |
poly |
|
face (in 2D), poly (in 3D) |
|
|
simulation step |
ver, edge, face, poly |
Variables consisting of several values (
vers, etc.) are not available for sorting (option-sort).For a cell, the
bodyvariable is defined as follows:
In the general case (
body, no argument provided), it is an integer equal to0if the cell is at the domain boundary, i.e. if it shares at least one face with it (edge in 2D), and is equal to1or higher otherwise. This is determined as follows: if a cell is surrounded by cells withbodyvalues equal to or higher thann, itsbodyvalue is equal ton + 1. Therefore,bodytends to increase with the distance to the domain boundary and can be used to define cells that may suffer from boundary effects.In the case where an expression is provided as argument (
body(<expr>)), the expression is a logical expression that defines the boundary to consider, from the domain face (edge in 2D) labels (for a cube,x0,x1,y0,y1,z0andz1). For example,body(z0||z1)considers only thez0andz1domain faces as the boundary, and the more exoticbody(x1&&y0||z1)considers only the intersection between thex1andy0domain faces, and thez1domain face as the boundary.For entities of lower dimension than cells (vertices, edges and faces),
bodyis equal to the maximumbodyvalue of the cells they belong to.
Raster Tessellation Keys
Available keys for raster tessellation itself are provided below.
Key |
Descriptor |
Apply to |
|
dimension |
tesr |
|
number of voxels in direction x |
tesr |
|
number of voxels in direction y |
tesr |
|
number of voxels in direction z |
tesr |
|
number of voxels in total |
tesr |
|
origin x coordinate |
tesr |
|
origin y coordinate |
tesr |
|
origin z coordinate |
tesr |
|
voxel size in direction x |
tesr |
|
voxel size in direction y |
tesr |
|
voxel size in direction z |
tesr |
|
raster size in direction x |
tesr |
|
raster size in direction y |
tesr |
|
raster size in direction z |
tesr |
|
raster size (surface area/volume in 2D/3D) |
tesr |
|
surface area |
tesr |
|
volume |
tesr |
|
size (surface area/volume in 2D/3D) |
tesr |
|
x coordinate |
tesr |
|
y coordinate |
tesr |
|
z coordinate |
tesr |
|
x, y and z coordinates |
tesr |
|
simulation step |
tesr |
Available keys for raster tessellation seeds, cells, cell groups and voxels are provided below. Mathematical and logical expressions based on these keys can also be used. To turn a key value into a value relative to the mean over all entities (e.g.the relative cell size), append the key expression with the :rel modifier. To turn a key value into a value which holds for a unit cell size, append the key expression with the :uc modifier.
General
Key |
Descriptor |
Applies to |
|
identifier |
seed, cell, group, voxel |
|
cell |
voxel |
|
orientation is defined |
voxel |
|
Laguerre weight |
seed |
|
simulation step |
tesr |
Geometry
Key |
Descriptor |
Applies to |
|
x coordinate |
seed, cell, voxel |
|
y coordinate |
seed, cell, voxel |
|
z coordinate |
seed, cell, voxel |
|
x, y and z coordinates |
seed, cell, voxel |
|
x coordinate (in voxel) |
voxel |
|
y coordinate (in voxel) |
voxel |
|
z coordinate (in voxel) |
voxel |
|
x, y and z coordinates (in voxel) |
voxel |
|
minimum x coordinate (in voxel) |
cell |
|
minimum y coordinate (in voxel) |
cell |
|
minimum z coordinate (in voxel) |
cell |
|
maximum x coordinate (in voxel) |
cell |
|
maximum y coordinate (in voxel) |
cell |
|
maximum z coordinate (in voxel) |
cell |
|
domain minimum x coordinate (in voxel), always 1 |
domain |
|
domain minimum y coordinate (in voxel), always 1 |
domain |
|
domain minimum z coordinate (in voxel), always 1 |
domain |
|
domain maximum x coordinate (in voxel) |
domain |
|
domain maximum y coordinate (in voxel) |
domain |
|
domain maximum z coordinate (in voxel) |
domain |
|
surface area |
cell, group (in 2D) |
|
volume |
cell, group (in 3D) |
|
size (surface area/volume in 2D/3D) |
cell, group |
|
number of voxels |
cell |
|
surface area fraction |
group (in 2D) |
|
volume fraction |
group (in 3D) |
|
size fraction (surface area/volume fraction in 2D/3D) |
group |
|
equivalent diameter [4] |
cell |
|
equivalent radius |
cell |
|
convexity [7] |
cell |
Orientation
Key |
Descriptor |
Applies to |
|
voxel, cell |
|
|
grain orientation spread [8] |
cell |
|
orientation distribution anisotropy / principal angles [13] |
cell |
|
orientation distribution anisotropy / principal axes [13] |
cell |
|
orientation distribution anisotropy factor [13] |
cell |
|
orientation distribution anisotropy / principal delta angles [14] |
cell |
Tessellation Optimization Keys
Time Keys
The available keys for option -morphooptilogtime are provided below. Use iter(<factor>), where factor is an integer reduction factor, to log values only at specific iteration numbers.
Key |
Descriptor |
|
iteration number |
|
number of updated variables |
|
number of updated seeds |
|
list of updated seeds |
|
number of updated cells |
|
list of updated cells |
|
time for variable update |
|
time for seed update |
|
time for cell update initialization |
|
time for cell update kdtree computation |
|
time for cell update shift computation |
|
time for cell update neighbor computation |
|
time for cell update cell computation |
|
time for cell update others |
|
total time for cell update |
|
time for (objective function) value update |
|
time for (objective function) value update / initialization |
|
time for (objective function) value update / penalty computation |
|
time for (objective function) value update / value computation |
|
time for (objective function) value update / value computation / cell values |
|
time for (objective function) value update / value computation / computation |
|
time for (objective function) value update / computation |
|
total time |
|
cumulative total time |
Variable Keys
The available keys for option -morphooptilogvar are provided below. Use iter(<factor>), where factor is an integer reduction factor, to log values only at specific iteration numbers.
Key |
Descriptor |
Apply to |
|
iteration number |
n/a |
|
identifier |
seed |
|
x coordinate |
seed |
|
y coordinate |
seed |
|
z coordinate |
seed |
|
weight |
seed |
Objective Function Value Keys
The available keys for option -morphooptilogval are provided below. Use iter(<factor>), where factor is an integer reduction factor, to log values only at specific iteration numbers.
Key |
Descriptor |
|
iteration number |
|
value |
|
minimal value |
|
value, without smoothing |
|
minimal value, without smoothing |
|
|
|
|
|
error on the objective function (see |
|
relative error on the objective function (see |
|
optimization loop |
|
current plateau length [15] |
Statistical Distribution Keys
The available keys for option -morphooptilogdis are provided below. PDF stands for probability density function and CDF stands for cumulative probability density function. Use iter(<factor>), where factor is a reduction factor, to log values only at specific iteration numbers.
Key |
Descriptor |
|
iteration number |
|
x coordinate |
|
target PDF |
|
target CDF |
|
current PDF |
|
current CDF |
|
target PDF, not smoothed |
|
target CDF, not smoothed |
|
current CDF, not smoothed |
Raster Tessellation Voxel Keys
The available keys for option -morphooptilogtesr are provided below. Values are written for each voxel used to compute the objective function. Use iter(<factor>), where factor is a reduction factor, to log values only at specific iteration numbers.
Key |
Descriptor |
|
iteration number |
|
cell identifier |
|
x coordinate |
|
y coordinate |
|
z coordinate |
|
distance to the cell |
Orientation Optimization Keys
Variable Keys
The available keys for option -orioptilogvar are provided below. For all orientation descriptors but quaternion, the returned orientation are located in the fundamental region. Use iter(<factor>), where factor is an integer reduction factor, to log values only at specific iteration numbers.
Key |
Descriptor |
Apply to |
|
iteration number |
n/a |
|
identifier |
seed |
|
Rodrigues vector |
seed |
|
Euler angles (Bunge convention) |
seed |
|
Euler angles (Kocks convention) |
seed |
|
Euler angles (Roe convention) |
seed |
|
Rotation matrix |
seed |
|
rotation axis / angle pair |
seed |
|
quaternion |
seed |
Mesh Keys
Available keys for a mesh itself are provided below. “co” stands for “cohesive”.
Key |
Descriptor |
Apply to |
|
element number |
{0-3}D,co mesh |
|
node number |
{0-3}D mesh |
|
elset number |
{0-3}D,co mesh |
|
partition number |
highest-dimension mesh |
|
x coordinate |
{0-3}D mesh |
|
y coordinate |
{0-3}D mesh |
|
z coordinate |
{0-3}D mesh |
|
x, y and z coordinates |
{0-3}D mesh |
|
length |
1D mesh |
|
surface area |
2D mesh |
|
volume |
3D mesh |
|
size (length/area/volume in 1D/2D/3D) |
{1-3}D mesh |
|
simulation step |
{0-3}D,co mesh |
Available keys for mesh node, elements and element sets (of all dimensions) and points are provided below. “co” stands for “cohesive”.
Key |
Descriptor |
Apply to |
|
identifier |
node, {0-3}D,co elt, {0-3}D,co elset |
|
x coordinate |
node, {0-3}D,co elt, {0-3}D elset |
|
y coordinate |
node, {0-3}D,co elt, {0-3}D elset |
|
z coordinate |
node, {0-3}D,co elt, {0-3}D elset |
|
x, y and z coordinates |
node, {0-3}D,co elt, {0-3}D elset |
|
lowest parent elt dimension |
node |
|
0D elset |
0D elt |
|
1D elset |
1D elt |
|
2D elset |
2D elt |
|
3D elset |
3D elt |
|
Cohesive elset |
co elt |
|
partition |
{0-3}D elt, node |
|
group |
{0-3}D elt, {0-3}D elset |
|
identifier of the corresponding tess cell at scale |
2D elset, 3D elset |
|
scale |
{0-2}D elset [3] |
|
cylinder polygonization [19] |
1D elt, 1D elset |
|
volume |
3D elt, 3D elset |
|
volume, orientation-space-wise [20] |
3D elt |
|
surface area |
2D elt |
|
equivalent diameter |
{2,3}D elt, {2,3}D elset |
|
equivalent radius |
{2,3}D elt, {2,3}D elset |
|
average edge length |
{0-3}D elt, 1D elset |
|
edge lengths |
2D elt, 3D elt |
|
elset volume |
3D elt |
|
elset area |
2D elt |
|
elset length |
1D elt |
|
radius ratio |
3D elt |
|
average radius ratio |
3D elset |
|
min radius ratio |
3D elset |
|
max radius ratio |
3D elset |
|
Osize |
3D elset |
|
number of elements |
{0-3}D,co elset |
|
elements |
{0-3}D,co elset |
|
number of nodes |
{0-3}D,co elset |
|
nodes |
{0-3}D,co elset |
|
body level |
{0-3}D elt, {0-3}D elset |
|
body level, relative to the elset boundary |
{1-3}D elt |
|
type of domain [16] |
{0-2}D elt, {0-2}D elset |
|
closest point of the 2D mesh |
node, 3D elt |
|
distance to |
node, 3D elt |
|
vector to |
node, 3D elt |
|
outgoing normal vector at |
node, 3D elt |
|
periodic (1 if periodic, 0 otherwise) |
{0,1}D elt, 2D elt (in 3D), {0,1}D elset, 2D elset (in 3D) |
|
color in Rodrigues vector convention [17] |
node |
|
color in IPF convention, cubic symmetry [18] |
node |
|
color in IPF convention, hexagonal symmetry [18] |
node |
|
1 if in orientation fiber and 0 otherwise |
3D elt, 3D elset |
|
disorientation angle (in degrees) |
1D elt and elset (in 2D), 2D elt and elset (in 3D) |
|
grain orientation spread [8] |
{2,3}D elset |
|
grain orientation spread estimated from the orientation distribution |
[8] {2,3}D elset |
|
2D elt (in 2D), 2D elset (in 2D), 3D elt (in 3D), 3D elset (in 3D) |
|
|
simulation step |
{0-3}D,co mesh |
Variables beginning with 2dmesh are only available for statistics (options beginning with -stat of module -M); for elements, they apply to the centroids.
Point Keys
Available keys for points are provided below.
Key |
Descriptor |
Apply to |
Require |
|
identifier |
point |
|
|
x coordinate |
point |
|
|
y coordinate |
point |
|
|
z coordinate |
point |
|
|
cell |
point |
tessellation |
|
containing element |
point |
mesh |
|
containing elset |
point |
mesh |
|
coordinates of the closest point of the 2D mesh |
point |
3D mesh |
|
distance to |
point |
3D mesh |
|
vector to |
point |
3D mesh |
|
outgoing normal vector of the 2D mesh at |
point |
3D mesh |
Simulation Results
A result of a Simulation Directory (.sim) can be invoked simply from its name. A component of a vectorial or tensorial result can be invoked by prefixing the component to the name, as in coo1, stress11, etc. For a symmetrical tensor (for which only 6 values are stored), t, both t<i><j> and t<j><i> are valid. The type of a result of the simulation directory is determined automatically. Tessellation results can be obtained from the cell results, by averaging or other statistical treatments. Similarly, elset and mesh results can be obtained from the element results, by averaging or other statistical treatments.
Available results / keys for nodes are the following:
Key |
Descriptor |
Apply to |
|
displacement (computed from positions) |
node |
Available results / keys for elements sets are the following:
Key |
Descriptor |
Apply to |
|
average orientation |
elset, mesh |
|
grain orientation spread [8] |
elset |
|
grain orientation spread computed from |
elset |
|
orientation distribution principal angles |
elset, mesh |
|
orientation distribution principal axes |
elset, mesh |
|
orientation distribution factor |
elset, mesh |
|
ODF defined at elements of orientation space (see also below) |
tess, tesr, mesh, cell, elt, elset |
|
ODF defined at nodes of orientation space (see also below) |
tess, tesr, mesh |
|
|
mesh |
|
|
mesh |
The ODF (odf or odfn) of a tessellation or mesh is computed over orientation space (provided using -orispace) from the orientations of the (tessellation) cells or (mesh) elsets. The (optional) parameters are:
input: the input used for the orientations, eitherelsetsoreltsfor a mesh (defaultelsets);theta: the standard deviation of the kernel (in degrees);weight: the weight of a cell or elset, which can be a real value or an expression based on the Tessellation Keys (for cells) or Mesh Keys (for elsets) – by default, the volumes of the cells or elsets are used;clustering: a logical value controlling orientation clustering, which can be0(for no clustering) or1(for clustering); the default is0for cells or elsets and1for voxels or elements;cutoff: the cut-off factor used to compute the ODF, which can beall(for no cut-off) or any positive real value (default5).
For a cell, element or elset, odf returns the value of the ODF of the tessellation or mesh at the corresponding orientation (and simulation step).
The orifield and orifieldn of a mesh is computed over orientation space (provided using -orispace) from the values of the (mesh) elsets. The mandatory parameter is:
var: the variable, which must be defined for elsets (i.e., have its files in the simulation directory);
and the optional parameters are:
theta: the standard deviation of the kernel (in degrees);weight: the weight of an elset, which can be a real value or an expression based on the Mesh Keys (for elsets) – by default, the volumes of the elsets are used.
Rotations and Orientations
Rotation and Orientation Descriptors
Rotations and orientations can be described using the following descriptors.
Key |
Descriptor |
Number of parameters |
|
Rodrigues vector |
3 |
|
Euler angles (Bunge convention) |
3 |
|
Euler angles (Kocks convention) |
3 |
|
Euler angles (Roe convention) |
3 |
|
rotation matrix |
9 |
|
rotation axis / angle pair |
4 |
|
quaternion |
4 |
The convention can be added to the descriptor, either active or passive, as in rodrigues:active. When no convention is provided, passive is assumed.
Some options can take parameter values as argument, in which case the orientation must be expressed as <descriptor>(<parameter1>,<parameters2>,...). An example is rodrigues(0.1,0.2,0.3).
Orientation Convention
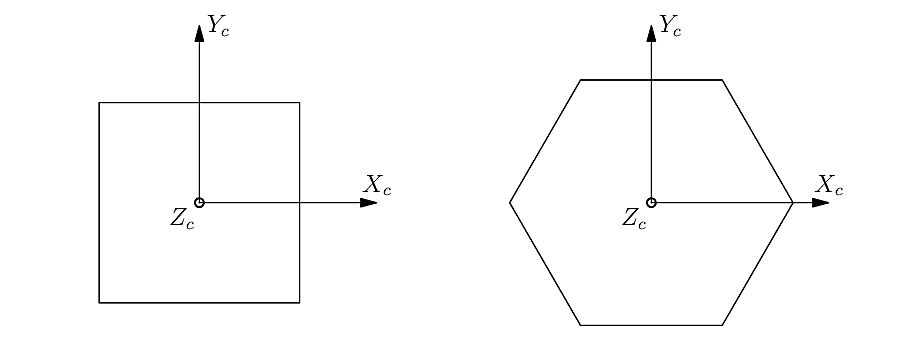
The crystal coordinate systems are attached to the crystal lattice as illustrated below, in the case of cubic and hexagonal symmetries:
The so-called “passive” orientation convention is used by default, which is based on the rotation of the sample coordinate system into the crystal coordinate system. Under this convention, the values of all Rotation and Orientation Descriptors are provided below for a simple (but representative) configuration that corresponds to a rotation of 30° about \(X_s\):
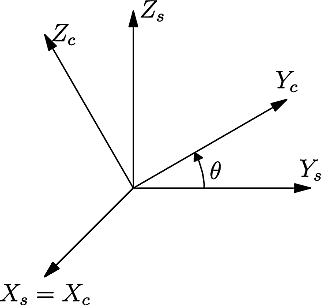
Descriptor |
Value (angles in degrees) |
Rodrigues vector |
\((0.267949192,\,0,\,0)\) |
Euler angles (Bunge convention) |
\((0,\,30,\,0)\) |
Euler angles (Kocks convention) |
\((270,\,30,\,90)\) |
Euler angles (Roe convention) |
\((270,\,30,\,90)\) |
rotation matrix |
\(\left(\begin{array}{ccc}1 & 0 & 0 \\ 0 & 0.866025404 & 0.5 \\ 0 & -0.5 & 0.866025404\\\end{array}\right)\) |
rotation axis / angle pair |
\((1,\,0,\,0) / 30\) |
quaternion |
\((0.965925826,\,0.258819045,\,0,\,0)\) |
The values of the orientation descriptors under the “active” convention are obtained by taking the opposite rotation.
Ideal Orientations
Keys are available for ideal orientations (lowercased is accepted):
Key |
Miller indices |
|
\((0\,0\,1)[1\,0\,0]\) |
|
\((0\,1\,1)[1\,0\,0]\) |
|
\((1\,0\,1)[\overline{1}\,0\,1]\) |
|
\((0\,0\,1)[1\,\overline{1}\,0]\) |
|
\((1\,2\,3)[6\,3\,\overline{4}]\) |
|
\((\overline{1}\,2\,3)[6\,\overline{3}\,4]\) |
|
\((1\,\overline{2}\,3)[6\,\overline{3}\,\overline{4}]\) |
|
\((\overline{1}\,\overline{2}\,3)[6\,3\,4]\) |
|
\((1\,1\,0)[1\,\overline{1}\,2]\) |
|
\((\overline{1}\,1\,0)[1\,1\,\overline{2}]\) |
|
\((1\,1\,2)[1\,1\,\overline{1}]\) |
|
\((\overline{1}\,1\,2)[1\,\overline{1}\,1]\) |
When loading orientations from an external file, use file(<file_name>[,des=<descriptor>]) where the orientation descriptor is among those listed above and is rodrigues:passive by default.
Orientation Fibers
Orientation fibers are defined by a crystal direction being parallel to a sample direction. Depending on the context, an angular tolerance or distribution with respect to the theoretical fiber can also be defined:
fiber(<dirc_x>,<dirc_y>,<dirc_z>,<dirs_x>,<dirs_y>,<dirs_z>), where (<dirc_x>,<dirc_y>,<dirc_z>) is the crystal direction and (<dirs_x>,<dirs_y>,<dirs_z>) is the sample direction, corresponds to an ideal orientation fiber;fiber(<dirc_x>,<dirc_y>,<dirc_z>,<dirs_x>,<dirs_y>,<dirs_z>,<theta>), where<theta>is an angle expressed in degrees, corresponds to an orientation fiber with the angular tolerance<theta>from the ideal fiber;fiber(<dirc_x>,<dirc_y>,<dirc_z>,<dirs_x>,<dirs_y>,<dirs_z>):normal(<var>=<val>), where<var>can bethetaorthetam) and<val>is the value, corresponds to an orientation fiber with a normal (Gaussian) disorientation normal to the ideal fiber.
Crystal Symmetries
Crystal symmetries can be described using the following descriptors.
Key |
Descriptor |
Number of operators |
|
triclinic (Laue group \(\overline{1}\)) |
24 |
|
cubic |
24 |
|
hexagonal |
1 |
|
Laue group \(\overline{1}\) |
1 |
|
Laue group \(2/m\) |
2 |
|
Laue group \(mmm\) |
4 |
|
Laue group \(4/m\) |
4 |
|
Laue group \(4/mmm\) |
8 |
|
Laue group \(\overline{3}\) |
3 |
|
Laue group \(\overline{3}m\) |
6 |
|
Laue group \(6/m\) |
6 |
|
Laue group \(6/mmm\) |
12 |
|
Laue group \(m\overline{3}\) |
12 |
|
Laue group \(m\overline{3}m\) |
24 |
Colors and Color Maps
Colors
The available colors are provided below, with their corresponding RGB channel values (ranging from 0 to 255). Any other color can be defined from the RGB channel values, under format <R_value>:<G_value>:<B_value>.
Key |
RGB value |
|
(0, 0, 0) |
|
(255, 0, 0) |
|
(0, 255, 0) |
|
(0, 0, 255) |
|
(255, 255, 0) |
|
(255, 0, 255) |
|
(0, 255, 255) |
|
(255, 255, 255) |
|
(128, 0, 0) |
|
(0, 0, 128) |
|
(127, 255, 0) |
|
(0, 255, 127) |
|
(128, 128, 0) |
|
(128, 0, 128) |
|
(0, 128, 128) |
|
(128, 128, 128) |
|
(0, 191, 255) |
|
(124, 252, 0) |
|
(64, 64, 64) |
|
(255, 69, 0) |
|
(192, 192, 192) |
|
(255, 250, 250) |
|
(139, 0, 0) |
|
(0, 0, 139) |
|
(255, 140, 0) |
|
(240, 255, 255) |
|
(248, 248, 255) |
|
(255, 255, 240) |
|
(0, 0, 205) |
|
(255, 182, 193) |
|
(245, 255, 250) |
|
(75, 0, 130) |
|
(240, 128, 128) |
|
(255, 192, 203) |
|
(255, 127, 80) |
|
(250, 128, 114) |
|
(255, 250, 240) |
|
(127, 255, 212) |
|
(255, 250, 205) |
|
(255, 215, 0) |
|
(0, 100, 0) |
|
(255, 165, 0) |
|
(240, 248, 255) |
|
(224, 255, 255) |
|
(255, 255, 224) |
|
(139, 0, 139) |
|
(0, 139, 139) |
|
(205, 133, 63) |
|
(70, 130, 180) |
|
(255, 240, 245) |
|
(255, 245, 238) |
|
(0, 250, 154) |
|
(72, 61, 139) |
|
(184, 134, 11) |
|
(255, 160, 122) |
|
(255, 228, 196) |
|
(135, 206, 250) |
|
(250, 250, 210) |
|
(240, 255, 240) |
|
(255, 248, 220) |
|
(255, 218, 185) |
|
(245, 245, 245) |
|
(255, 99, 71) |
|
(112, 128, 144) |
|
(255, 105, 180) |
|
(253, 245, 230) |
|
(255, 235, 205) |
|
(189, 183, 107) |
|
(255, 228, 181) |
|
(0, 206, 209) |
|
(60, 179, 113) |
|
(199, 21, 133) |
|
(238, 130, 238) |
|
(173, 255, 47) |
|
(255, 239, 213) |
|
(143, 188, 143) |
|
(188, 143, 143) |
|
(255, 20, 147) |
|
(139, 69, 19) |
|
(148, 0, 211) |
|
(30, 144, 255) |
|
(119, 136, 153) |
|
(222, 184, 135) |
|
(255, 222, 173) |
|
(250, 240, 230) |
|
(123, 104, 238) |
|
(64, 224, 208) |
|
(135, 206, 235) |
|
(72, 209, 204) |
|
(245, 245, 220) |
|
(255, 228, 225) |
|
(210, 180, 140) |
|
(250, 235, 215) |
|
(216, 191, 216) |
|
(50, 205, 50) |
|
(233, 150, 122) |
|
(176, 196, 222) |
|
(65, 105, 225) |
|
(152, 251, 152) |
|
(220, 20, 60) |
|
(245, 222, 179) |
|
(186, 85, 211) |
|
(230, 230, 250) |
|
(240, 230, 140) |
|
(144, 238, 144) |
|
(175, 238, 238) |
|
(47, 79, 79) |
|
(153, 50, 204) |
|
(46, 139, 87) |
|
(154, 205, 50) |
|
(138, 43, 226) |
|
(219, 112, 147) |
|
(107, 142, 35) |
|
(147, 112, 219) |
|
(244, 164, 96) |
|
(85, 107, 47) |
|
(102, 205, 170) |
|
(106, 90, 205) |
|
(238, 232, 170) |
|
(34, 139, 34) |
|
(25, 25, 112) |
|
(32, 178, 170) |
|
(211, 211, 211) |
|
(218, 112, 214) |
|
(100, 149, 237) |
|
(160, 82, 45) |
|
(178, 34, 34) |
|
(176, 224, 230) |
|
(205, 92, 92) |
|
(105, 105, 105) |
|
(173, 216, 230) |
|
(210, 105, 30) |
|
(165, 42, 42) |
|
(218, 165, 32) |
|
(220, 220, 220) |
|
(221, 160, 221) |
|
(95, 158, 160) |
Color Maps
Color Map for Integer Values
The color map or palette used to represent integer values is defined from the above color list, by excluding colors of brightness below 0.2 and above 0.8. The brightness is defined as the average of the channel values divided by 255. The resulting list of colors is: (1) red, (2) green, (3) blue, (4) yellow, (5) magenta, (6) cyan, (7) chartreuse, (8) springgreen, (9) olive, (10) purple, (11) teal, (12) gray, (13) deepskyblue, (14) lawngreen, (15) darkgray, (16) orangered, (17) silver, (18) darkorange, (19) mediumblue, (20) indigo, (21) lightcoral, (22) coral, (23) salmon, (24) aquamarine, (25) gold, (26) orange, (27) darkmagenta, (28) darkcyan, (29) peru, (30) steelblue, (31) mediumspringgreen, (32) darkslateblue, (33) darkgoldenrod, (34) lightsalmon, (35) lightskyblue, (36) tomato, (37) slategray, (38) hotpink, (39) darkkhaki, (40) darkturquoise, (41) mediumseagreen, (42) mediumvioletred, (43) violet, (44) greenyellow, (45) darkseagreen, (46) rosybrown, (47) deeppink, (48) saddlebrown, (49) darkviolet, (50) dodgerblue, (51) lightslategray, (52) burlywood, (53) mediumslateblue, (54) turquoise, (55) skyblue, (56) mediumturquoise, (57) tan, (58) limegreen, (59) darksalmon, (60) lightsteelblue, (61) royalblue, (62) palegreen, (63) crimson, (64) mediumorchid, (65) khaki, (66) lightgreen, (67) darkslategray, (68) darkorchid, (69) seagreen, (70) yellowgreen, (71) blueviolet, (72) palevioletred, (73) olivedrab, (74) mediumpurple, (75) sandybrown, (76) darkolivegreen, (77) mediumaquamarine, (78) slateblue, (79) forestgreen, (80) midnightblue, (81) lightseagreen, (82) orchid, (83) cornflowerblue, (84) sienna, (85) firebrick, (86) indianred, (87) dimgray, (88) chocolate, (89) brown, (90) goldenrod, (91) plum and (92) cadetblue.
Color Maps for Real Values
The color map used to represent real values is smooth and obtained by interpolation between nominal colors. Tinycolormap is used to generate standard color maps, and the default is viridis. The color maps are
Key |
Color bar |
viridis |

|
cividis |

|
magma |

|
inferno |

|
plasma |

|
parula |

|
heat |

|
hot |

|
jet |

|
gray |

|
github |

|
Alternatively, a custom color map can be provided as custom(<color1>,<color2>,...). Neper’s legacy color map (version \(< 4\)) is custom(blue,cyan,yellow,green) and can also be obtained using legacy:
legacy |

|
Finally, it is possible to gradually fade the start of a color map, to make it starts with white. This can be done using the fade modifier, following the syntax <colormap>:fade[(threshold)]. The threshold ranges from 0 to 1 and is equal to 0.1 by default. Fading is applied linearly from 0 (full fading) to the threshold (no fading). Examples are show below:
viridis:fade |

|
viridis:fade(0.2) |

|